iFluor® 860 maleimide
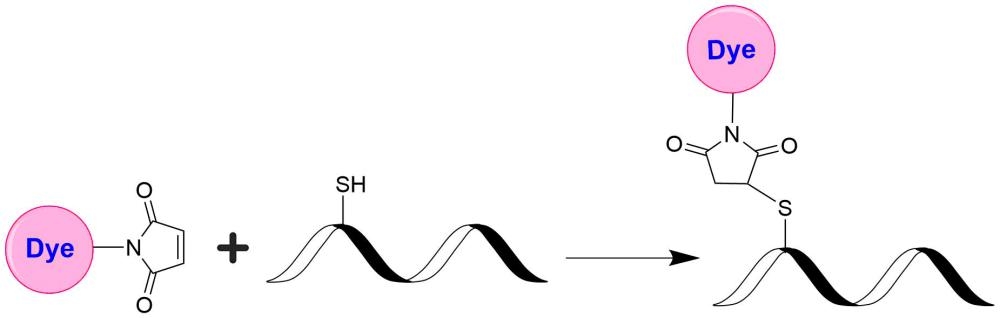
In vivo fluorescence imaging uses a sensitive camera to detect the fluorescence emission from fluorophores in whole-body living small animals. To overcome the photon attenuation in living tissue, fluorophores with long emission at the infrared (IR) region are generally preferred. Recent advances in imaging strategies and reporter techniques for in vivo fluorescence imaging include novel approaches to improve the specificity and affinity of the probes and to modulate and amplify the signal at target sites for enhanced sensitivity. Further emerging developments aim to achieve high-resolution, multimodality, and lifetime-based in vivo fluorescence imaging. Our iFluor® 860 is designed to label proteins and other biomolecules with infrared fluorescence. Conjugates prepared with iFluor® 860 have excitation and emission in the IR range. iFluor® 860 dye emission is well separated from commonly used far-red fluorophores such as Cy5, Cy7, or allophycocyanin (APC), facilitating multicolor analysis. This fluorophore is also useful for small animal in-vivo imaging applications or other imaging applications requiring IR detection. iFluor® 860 maleimide is thiol-reactive and can be readily used to conjugate thiol-containing biomolecules.

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 1408 | 1 mg | Price |
Physical properties
| Molecular weight | 1647.66 |
| Solvent | DMSO |
Spectral properties
| Correction factor (260 nm) | 0.1 |
| Correction factor (280 nm) | 0.14 |
| Extinction coefficient (cm -1 M -1) | 250000 1 |
| Excitation (nm) | 853 |
| Emission (nm) | 878 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 27, 2026

