iFluor® 488 maleimide
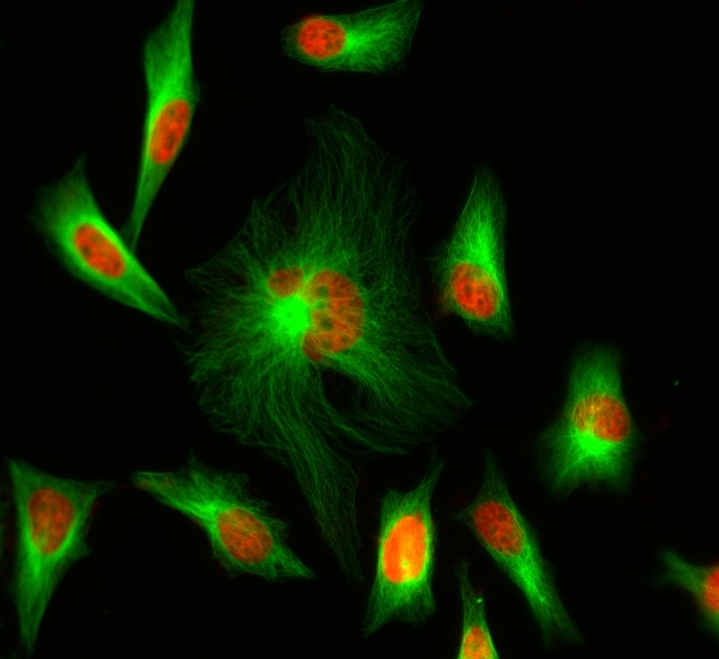
Although FITC is still the most popular fluorescent labeling dye for preparing green fluorescent bioconjugates, there are certain limitations with FITC, such as severe photobleaching for microscope imaging and pH-sensitive fluorescence. Protein conjugates prepared with iFluor® 488 dyes are far superior to conjugates of fluorescein derivatives such as FITC. iFluor® 488 conjugates are significantly brighter than fluorescein conjugates and are much more photostable. Additionally, the fluorescence of iFluor® 488 is not affected by pH (4-10). This pH insensitivity is a major improvement over fluorescein, which emits its maximum fluorescence only at pH above 9. iFluor® 488 maleimide is reasonably stable and shows good reactivity and selectivity with the thiol group. This iFluor® 488 has spectral properties and reactivity similar to Alexa Fluor® 488 maleimide ( Alexa Fluor® is the trademark of Invitrogen).

| Catalog | Size | Price | Quantity |
|---|---|---|---|
| 1062 | 1 mg | Price |
Physical properties
| Molecular weight | 881.76 |
| Solvent | DMSO |
Spectral properties
| Correction factor (260 nm) | 0.21 |
| Correction factor (280 nm) | 0.11 |
| Extinction coefficient (cm -1 M -1) | 75000 1 |
| Excitation (nm) | 491 |
| Emission (nm) | 516 |
| Quantum yield | 0.9 1 |
Storage, safety and handling
| H-phrase | H303, H313, H333 |
| Hazard symbol | XN |
| Intended use | Research Use Only (RUO) |
| R-phrase | R20, R21, R22 |
| Storage | Freeze (< -15 °C); Minimize light exposure |
| UNSPSC | 12171501 |
Contact us
| Telephone | |
| Fax | |
| sales@aatbio.com | |
| International | See distributors |
| Bulk request | Inquire |
| Custom size | Inquire |
| Technical Support | Contact us |
| Request quotation | Request |
| Purchase order | Send to sales@aatbio.com |
| Shipping | Standard overnight for United States, inquire for international |
Page updated on February 27, 2026

