Affinity Purification
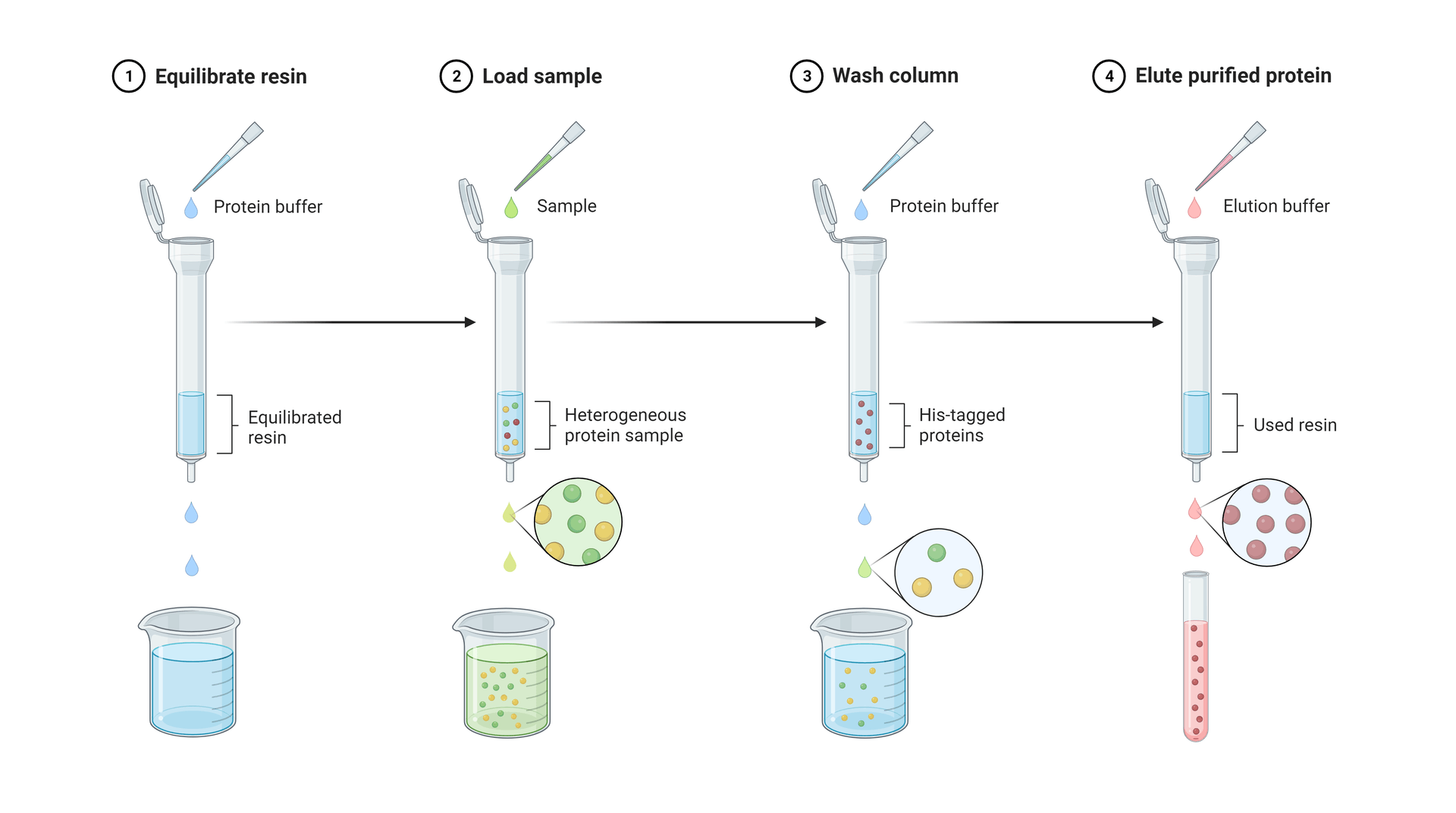
Simplified steps of an affinity purification procedure, in this case using a His-tag for the affinity interaction. Figure made in Biorender.
Table of Contents
Purified proteins can also be used in research or analytical purposes, including for assays on protein quantification, structural studies, identifying post-translational modifications and functional analysis. Affinity purification can offer much time saved in experimentation over less selective, multi-step procedures. In affinity purification native protein forms can also be identified apart from denatured forms in the same sample. While other purification methods separate molecules based on size (e.g., gel filtration, size-exclusion chromatography) or ionic interaction (e.g., ion exchange chromatography), affinity purification works by manipulating certain molecular properties and binding interactions between molecules. With the growing popularity of affinity purification, many materials and reagents are commercially available, come conveniently pre-prepped, and ready to use. Affinity purification can provide several hundred to thousand-fold levels of purification, but the success and resolution of the proteins depends on the method. Thus, it is important to optimize the affinity purification protocol to achieve the maximum capture and recovery of the target protein.
Purification Principle
There are multiple variations of affinity purification. In principle each work by manipulating the interaction strength differences between biomolecules between two elements, termed the mobile and stationary phases. In short, the stationary phase is either a solid complex or layer of a liquid adsorbed on the surface of the solid support. In affinity purification, the stationary phase normally includes specialized beads or affinity resin. The mobile phase is either a liquid or gaseous component that travels through the stationary phase and picks up the compounds to be tested along the way. The mobile phase is chosen according to the stationary phase, for example, if the stationary phase is polar, then the mobile phase has lower charge or is nonpolar.
In a standard experiment, the protein of interest is attached to a matrix of agarose beads or resin selectively bound by a ligand, often an antibody. The matrix contains a particular ligand, to which the molecule of interest has a specific binding affinity to. In this way the target molecule attracts and sticks to the matrix. Incubation with the sample and ligand resin in ideal conditions facilitates binding of the target protein to the matrix. After, unbound components of the sample are washed away from the matrix using a buffer. Elution can then release the purified protein by simply changing the buffer conditions, for example in a shift in pH, ionic strength, or molarity, or by using a competitive ligand. In this reversible interaction between the target proteins and the matrix, proteins can easily be collected in a purified, concentrated form
Table 1. Agarose Conjugates
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 55000 | Protein A-Agarose Resin | 1 mL |
| 55005 | Protein A-Agarose Resin | 5 mL |
| 55010 | Goat anti-mouse IgG (H&L) Agarose | 10 mg |
| 55015 | Goat anti-rabbit IgG (H&L) Agarose | 10 mg |
| 55016 | ReadiPrep™ Protein G-Agarose Resin | 1 mL |
| 55017 | ReadiPrep™ Protein G-Agarose Resin | 5 mL |
Affinity Chromatography
.png&w=1080&q=100)
Simplified affinity chromatography column interactions, from initial sample addition and incubation, through washing steps and eventual elution of purified target protein. Figure made in Biorender.
The experimental setup in affinity chromatography is also more complex and usually incorporates a pump, vacuum, or gravimetric system to force the mobile phase, buffers, and other reagents through the column. Affinity chromatography is especially useful to separate active biomolecules from denatured or functionally different forms. Additionally, this method can be used to isolate pure substances present at low concentration in large volumes of crude samples and also to remove specific contaminants.
Affinity chromatography comes with special elements compared to simpler forms of affinity purification. The chromatography matrix, also termed a column, is made of porous materials such as cellulose, agarose, polyacrylamide, polymethacrylate, or silica. Column selection should be based on the physical and chemical properties of the starting sample and reagents used. Column materials may also have pre-integrated functional groups with affinities for certain side chains on the target protein.
Table 2. ReadiUse™ Columns
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 60500 | ReadiUse™ Bio-Gel P-6 spin column | 5 Columns |
| 60504 | ReadiUse™ Disposable PD-10 Desalting Column | 5 Columns |
| 60505 | ReadiUse™ Disposable PD-10 Desalting Column | 10 Columns |
Affinity chromatography comes with special elements compared to simpler forms of affinity purification. The chromatography matrix, also termed a column, is made of porous materials such as cellulose, agarose, polyacrylamide, polymethacrylate, or silica. Column selection should be based on the physical and chemical properties of the starting sample and reagents used. Column materials may also have pre-integrated functional groups with affinities for certain side chains on the target protein.
Spacer arms can be incorporated into a setup to help enable better binding by overcoming any steric hindrance effects. These are typically a hydrocarbon chain, 4-12 atoms in length, that act as branched arms that attract and cling to target proteins on the ligand. Another critical consideration in affinity chromatography include ligands, which must be selected depending on the molecules to be separated. The ligands can be covalently immobilized or adsorbed on a solid surface via biochemical interactions, entrapped within a pore, or conjugated with a metal ion.
In a typical affinity chromatography protocol, the affinity column may first need to be briefly centrifuged to settle the resin at the bottom of the column, and wetted with a buffer to aid in ligand-protein binding. The crude sample mobile phase is then incubated with the stationary phase or affinity support. Ligands are immobilized to the solid support material by one of two ways:
Next, the column undergoes a series of washing steps where undesirable, non-target molecules are eluted using an appropriate buffer. Target molecules, in comparison, have a much stronger affinity for the stationary phase, so in these steps only the non-target molecules should be washed away. Lastly, the column goes through an elution step where target molecules are released from the ligands and collected, separately, as newly purified proteins. Experimental setup, binding, and elution conditions in affinity chromatography can vary greatly, so protocols should be optimized based on target protein.
Though many variations of the technique exist some common examples include DNA affinity chromatography, purification or removal of serine proteases, immunoaffinity, purification of recombinant proteins, lectin affinity chromatography, serum albumin purification, dye-ligand affinity chromatography, purification of biotin and biotinylated substances, and purification or removal of fibronectin.
Table 3. IDA ligands for Immobilized metal affinity chromatography (IMAC)
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 12630 | Biotin-X IDA | 1 mg |
| 12631 | IDA maleimide | 5 mg |
| 12632 | IDA succinimidyl ester | 5 mg |
In a typical affinity chromatography protocol, the affinity column may first need to be briefly centrifuged to settle the resin at the bottom of the column, and wetted with a buffer to aid in ligand-protein binding. The crude sample mobile phase is then incubated with the stationary phase or affinity support. Ligands are immobilized to the solid support material by one of two ways:
- Formation of covalent bonds between particular functional groups (e.g., primary amines, aldehydes, carboxylic acids, sulfhydryls) and the reactive groups
- Indirect coupling methods.
Next, the column undergoes a series of washing steps where undesirable, non-target molecules are eluted using an appropriate buffer. Target molecules, in comparison, have a much stronger affinity for the stationary phase, so in these steps only the non-target molecules should be washed away. Lastly, the column goes through an elution step where target molecules are released from the ligands and collected, separately, as newly purified proteins. Experimental setup, binding, and elution conditions in affinity chromatography can vary greatly, so protocols should be optimized based on target protein.
Though many variations of the technique exist some common examples include DNA affinity chromatography, purification or removal of serine proteases, immunoaffinity, purification of recombinant proteins, lectin affinity chromatography, serum albumin purification, dye-ligand affinity chromatography, purification of biotin and biotinylated substances, and purification or removal of fibronectin.
Spin Column and Batch Formats

Prepacked ReadiUse™ Bio-Gel P6 Spin Column. The column was packed with P-6 DG in PBS buffer for sample volume 50~100 uL. The MW limit is ~6000 and large molecules like proteins (MW > 15,000) will be eluted with a buffer.
The spin cup purification method provides improved efficiency of both the wash and elution steps in comparison to the batch method, and is appropriate for volumes ranging from approximately 50-300 µl of immobilized protein samples. The spin cup consists of two parts: the inner cup has a filter, typically made of paper, cellulose acetate or polyethylene, while the outer cup is a microcentrifuge tube. This apparatus allows the liquid fraction to be totally separated from the target protein-ligand sample upon centrifugation, and retained in the outer microcentrifuge tube. Many types of spin cups are commercially available with various pore sizes, volumes, and tube or cap configurations. Compared to affinity chromatography, washing and elution steps in the batch and spin cup formats are somewhat inefficient. Even so, these procedures use inexpensive materials, and offer quick preparation times along with easy experimental setup. More so, there exist many batch and spin cup kits for the purification of specific proteins, like cell surface proteins or glycoproteins, on the market.
In a typical protocol the sample and ligand-containing resin are initially mixed and allowed to interact. This incubation immobilizes the protein in a resin slurry, where the resin may contain just one ligand or multiple ligands if higher purification and recovery is desired. Next the immobilized protein-resin slurry is pipetted into either a microcentrifuge tube or spin cup column, for batch and spin cup formats, respectively. Then the sample is centrifuged at high speed for a short amount of time to separate the protein-resin slurry and liquid parts.
In the batch format, the supernatant must be carefully removed, and in the spin cup format the outer microcentrifuge may be discarded and replaced. Binding buffer is then added, the sample is resuspended and undergoes centrifugation, and the supernatant or liquid layer is removed again. The sample undergoes 4-5 rounds of resuspension and centrifugation to wash away and remove as much unbound non-target material as possible. Commonly, an additional salt wash with 1 M NaCl can also be included to help remove nonspecific material. After washing, an elution buffer is added to the microcentrifuge tube or spin column to resuspend the sample. After incubation, the sample is centrifuged and the supernatant is transferred to a separate microcentrifuge tube. This supernatant contains the newly unbound target proteins. 4-5 additional rounds of elution wash and centrifugation should be performed to ensure all target proteins from the sample are eluted.
Table 4. Ordering information for laboratory consumables and general laboratory equipment.
| Product ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| CytoCite™ BG100 Portable Fluorometer | 1 Fluorometer | CBG100 |
| CytoCite™ Sample Tube *500 µL for Qubit® and CycoCite™ fluorometers* | 500 Tubes | CCT100 |
| ReadiUse™ 10KD Spin Filter | 5 Filters | 60502 |
| ReadiUse™ Bio-Gel P-6 spin column | 5 Columns | 60500 |
| PD-10 Spin Adapter | 5 Adapters | 60506 |
| PD-10 Spin Adapter | 10 Adapters | 60507 |
| PD-10 Buffer Reservoir | 5 Pieces | 60508 |
| PD-10 Buffer Reservoir | 10 Pieces | 60509 |
Experimental Considerations
Materials, Reagents and Protein Selection
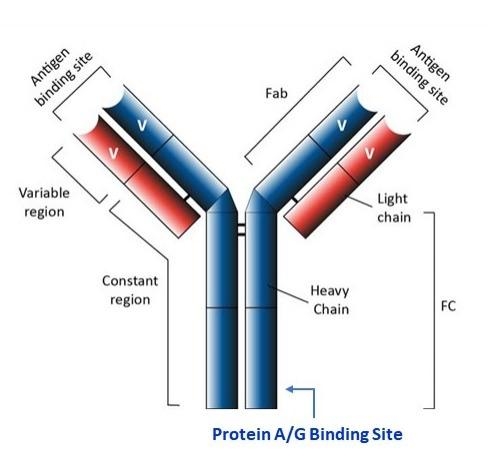
Protein A agarose beads have physical and chemical properties that enable them to be used in a variety of batch- or column-type affinity purification procedures. Protein A contains four high-affinity binding sites interacting with the Fc region of IgG-class antibodies from selected mammalian species.
As far as reagents go, the affinity medium should be thoroughly washed or filtered before use to remove all traces of storage solutions and preservatives. As good practice only high-quality water and chemicals should be used, and reusing affinity media should be avoided. Another initial step in designing an affinity purification study is in selecting proteins that will maximize target interactions. For example, if the target protein contains a zinc finger binding domain to attract DNA, then at least one other negative-control protein should be included that also contains the same binding domain. This addition will help identify proteins that uniquely interact with the target protein compared to other, general, zinc finger-binding proteins present.
The use of controls can also help improve the characterization of background associated with a given experimental protocol upon analysis. Positive controls should be a known protein that has already undergone purification and analysis, and that has a high-confidence set of interacting proteins identified by counterpart binding assays. Negative controls, on the other hand, should be any protein that is not expected to interact with the sample. For example, GFP is commonly chosen as a negative control since any interactions it induces are usually related to the epitope tag or resin-capture system.
| Datasets: | |
Affinity Tags and Ligands
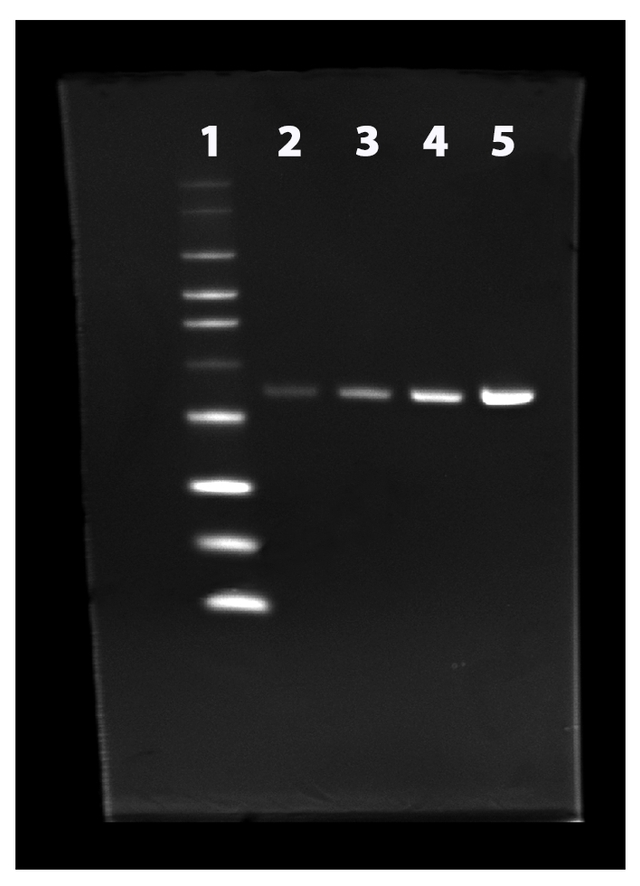
Two-fold dilution series of His-tagged annexin V were separated on a NuPAGE® 4-12% Bis-Tris gel and stained with the ProLite™ His-Tag Protein Gel Staining Kit according to standard protocols. Lane 1: His-tagged protein ladder, Lane 2 to 5: two-fold dilution of His-tagged annexin V.
Ligands selection key to the success of affinity purification, and the characteristics of a good ligand are that it can covalently attach to the matrix and not bind to non-target molecules for effective washing. Not only must it be able to retain its specific binding affinity for the target molecule, it must do so in a reversible manner for subsequent elution. Testing the affinity of the ligand to the target molecule may be a necessary initial step before purification since extremely low or high affinity can significantly reduce yield.
Table 5. 6xHIS Conjugates
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 12624 | 6XHis Succinimidyl Ester | 1 mg |
| 12626 | 6XHis maleimide | 1 mg |
| 12628 | 6XHis azide | 1 mg |
| 12629 | 6XHis alkyne | 1 mg |
| V102000 | Purified Mouse Anti-6xHIS Tag Antibody *Monoclonal* | 0.1 mg |
| V102005 | Purified Rabbit Anti-6xHIS Tag Antibody *Polyclonal* | 0.1 mg |
| V102010 | Biotin Rabbit Anti-6xHIS Tag Antibody *Polyclonal* | 0.1 mg |
| V102015 | FITC Rabbit Anti-6xHIS Tag Antibody *Polyclonal* | 0.1 mg |
| V102020 | HRP Rabbit Anti-6xHIS Tag Antibody *Polyclonal* | 0.1 mg |
Since ligand binding in affinity purification exploits spontaneous biological interactions, many successful target protein and ligand combinations have already been identified. For example enzymes are attracted by their specific substrate analogue, inhibitor, or cofactor, while antibodies are attracted by their distinct antigen, or virus. Lectins are drawn to a polysaccharide, glycoprotein, or cell surface receptor, while nucleic acids are drawn to their precise complementary base sequence, histone, nucleic acid polymerase, or nucleic acid binding protein. Additionally, hormones and vitamins will have high affinity for their unique cell receptor, or carrier protein, while metal ions have a high affinity for His-fusion proteins, native proteins with histidine, or molecules with cysteine or tryptophan residues.
Experimental steps
There are a number of steps that go into any affinity purification protocol and each, too, must be carefully considered. More stringent purification procedures generally require substantially more starting material and include strict wash steps. Such conditions may prohibit identifying transient interactions or those that occur with low-abundance proteins. If preferred, sample recovery can be improved and manipulation speed for capturing transient interactions may be increased in a number of ways, by using single-step purification procedure, or opting to use magnetic beads rather than agarose resins for faster wash times.
Including a step that involves cryogenic lysis of the pellet, or even incorporating an in-line chromatographic strategy may also speed up the process. All of these methods, however, have the potential to increase the degree to which non-specifically interacting proteins are represented in the final sample analyzed. For this reason, challenges in affinity purification are not only how to optimize methods to obtain highly pure proteins, but also in developing bioinformatic methods to differentiate true protein interactions from background interactions.
Note: It is important to note that adjusting the sample to the composition and pH of the binding buffer may improve the binding efficiency of the target protein.
Also, in affinity chromatography, a high flow rate may be better for samples with strong ligand-target molecule affinities while a lower flow rate may be better for samples with weaker affinities. Ensuring that all unbound materials have been thoroughly washed through the column before proceeding with elution will always improve the purity of the target molecule.
Additional Analytical Assays
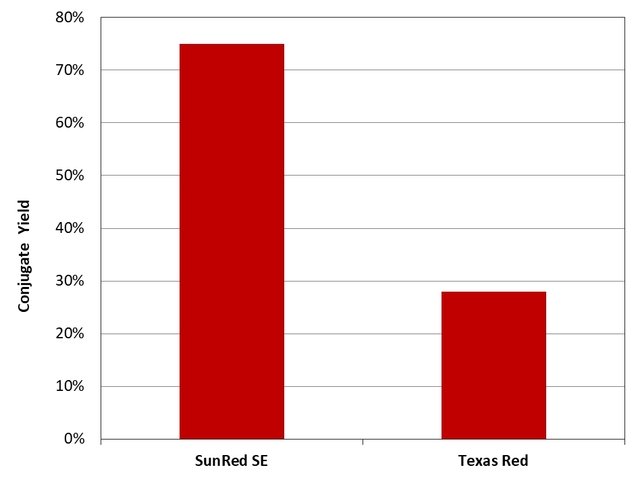
Texas Red® and SunRed™ SE were respectively mixed with Gly-gly-Ser-Ser-Arg-Trp in 1:1 molar ratio at pH=9.0 under the same conditions. The resulting conjugates were purified by HPLC using a reverse phase C18 column.
Western blots may also be useful to verify that epitope-tagged proteins are expressed and intact. Lowry or Bradford assays may also be used as secondary assays to determine total protein content. Alternatively, protein purity can be estimated by SDS-PAGE, isoelectric focusing, capillary electrophoresis, reversed phase chromatography or even mass spectrometry. ELISAs can be incorporated to focus on immune specific interactions to assess the active concentration of target molecules. If desired, SDS-PAGE, Western blot and ELISAs can be used to quantitate the genetically engineered molecules to which a specific tag has been attached. Many commercial kit assays also exist that have been specifically designed to detect a certain tag.
Removal of Contaminants
It is highly recommended to centrifuge and filter any sample immediately before chromatographic purification, as these steps help remove lipids, cell debris and particulate matter. Membrane filters that provide the least amount of nonspecific binding should be considered, like cellulose acetate or PVDF. Fractional precipitation may be incorporated to remove gross impurities from small sample volumes, and have occasionally been successfully employed in small-scale commercial production. This technique utilizes the principle of differential solubility, based on variance in hydrophobicity between proteins. The procedure employs increased salt concentrations to enhance hydrophobic interactions between the proteins and cause precipitation. A desalting step is also normally included in the methodology to remove persisting reagents and other contaminants. This step is crucial not only to ensure the purification of the product, but also to ensure the reaction between the ligands and target proteins terminates precisely when expected.
Table 6. Comparison of nitrocellulose and polyvinylidene fluoride membranes.
▲ ▼ | Nitrocellulose (NC) ▲ ▼ | Polyvinylidene Fluoride (PVDF) ▲ ▼ |
| Protein binding capacity | 80 to 100 µg/cm² | 170 to 200 µg/cm² |
| Binding interactions | Proteins bind to membrane through hydrophobic interactions | Proteins bind to membrane through hydrophobic and dipole interactions |
| Physical characteristics | Brittle, fragile and not chemically resistant | Physically durable and chemically resistant |
| Pore size | 0.1, 0.2 or 0.45µm | 0.1, 0.2 or 0.45µm |
| Membrane format | Pre-wetted with methanol | No need to pre-wet |
| Attributes |
|
|
In desalting, typically a mild reagent with a lower salt concentration is employed, like 25 mM NaCl, to prevent possible ionic interactions, though more volatile buffers, like 100 mM ammonium acetate or ammonium hydrogen carbonate, can be used if necessary. Desalting may be vital to remove remnants of other reagents used in cell preparation, like phenol red that has the capacity to bind and interact with some types of purification media.
Another traditional method of removing salt or other small molecules of a sample includes dialysis. Dialysis is generally very slow, requires large volumes of buffer, and presents the inherent risk of losing material due handling procedures, proteolytic breakdown, or non-specific binding to the dialysis membranes. Dialysis, however, is a good backup technique for the removal of contaminants when the sample or other reagents may react negatively in the presence of salts. It is important to remember that lipoproteins and other lipid material can rapidly clog chromatography columns, so it may be necessary to remove them before purification using precipitation agents like dextran sulfate or polyvinylpyrrolidone. Additionally some samples, like serum, can be filtered through glass wool to remove remaining lipids.
Product Ordering Information
Table 7. ReadiUse™ Spin Column and Adapter
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 60500 | ReadiUse™ Bio-Gel P-6 spin column | 5 Columns |
| 60506 | PD-10 Spin Adapter | 5 Pieces |
Table 8. ReadiUse™ Desalting Columns
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 60504 | ReadiUse™ Disposable PD-10 Desalting Column | 5 Columns |
| 60505 | ReadiUse™ Disposable PD-10 Desalting Column | 10 Columns |
Table 9. Ordering information for laboratory consumables and general laboratory equipment.
| Product ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| CytoCite™ BG100 Portable Fluorometer | 1 Fluorometer | CBG100 |
| CytoCite™ Sample Tube *500 µL for Qubit® and CycoCite™ fluorometers* | 500 Tubes | CCT100 |
| ReadiUse™ 10KD Spin Filter | 5 Filters | 60502 |
| ReadiUse™ Bio-Gel P-6 spin column | 5 Columns | 60500 |
| PD-10 Spin Adapter | 5 Adapters | 60506 |
| PD-10 Spin Adapter | 10 Adapters | 60507 |
| PD-10 Buffer Reservoir | 5 Pieces | 60508 |
| PD-10 Buffer Reservoir | 10 Pieces | 60509 |
Table 10. HIS Lite™ products
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 12610 | HIS Lite™ Cy3 Bis NTA-Ni Complex | 1 mg |
| 12613 | HIS Lite™ Cy5 Bis NTA-Ni Complex | 1 mg |
| 12615 | HIS Lite™ OG488-Tris NTA-Ni Complex | 100 µg |
| 12617 | HIS Lite™ iFluor® 568 Tris NTA-Ni Complex | 100 µg |
| 12618 | HIS Lite™ iFluor™ 647 Tris NTA-Ni Complex | 100 µg |
| 12619 | HIS Lite™ Cy5 Tris NTA-Ni Complex | 100 µg |
| 12620 | HIS Lite™ Cy3 Tris NTA-Ni Complex | 100 µg |
Table 11. Protein A for Affinity Purification
| Cat# ▲ ▼ | Product Name ▲ ▼ | Unit Size ▲ ▼ |
| 11041 | MegaWox™ polyHRP-Protein A Conjugate | 1 mg |
| 55000 | Protein A-Agarose Resin | 1 mL |
| 55005 | Protein A-Agarose Resin | 5 mL |
