Reverse Transcription PCR (RT-PCR)
Reverse transcription polymerase chain reaction (RT-PCR) is a core molecular technique designed to detect and quantitate total RNA or messenger RNA (mRNA), with a high degree of sensitivity and accuracy. In this modified version of the standard PCR process, mRNA, which serves as the initial template, is first reversed transcribed to complementary DNA (cDNA) and then subsequently amplified via PCR for downstream analysis. Relative to other techniques for measuring mRNA, such as Northern blot analysis, RNAse protection assays, or in situ hybridization, RT-PCR is significantly more robust at detecting the RNA transcript of any gene regardless of its relative abundance. Consequently, RT-PCR is widely used to quantitatively study gene expression, examine transcript variants, and generate cDNA templates for cloning and sequencing. Furthermore, RT-PCR has become an instrumental diagnostic tool for the detection of pathogens, including viruses that cause Ebola, HIV and the novel coronavirus disease (Covid-19).
Principles of Reverse Transcription PCR
In order to apply PCR to the study of RNA, the RNA sample must first undergo the process of reverse transcription to generate cDNA, using the enzyme reverse transcriptase. This RNA-dependent DNA polymerase, with the assistance of reverse transcription primers, deoxynucleotides (dNTPs: dATP, dTTP, dGTP and dCTP) and enzyme cofactors, catalyzes the synthesis of cDNA from its respective target RNA sequence. The resulting cDNA is then used as the template for PCR amplification, and depending upon the manner in which reverse transcription and PCR are executed this process can be classified as either one-step RT-PCR or two-step RT-PCR.
Video 1. RT-PCR animation. Animated video tutorial illustrating the key steps in reverse transcription polymerase chain reaction: (1) reverse transcription, (2) denaturation, (3) primer annealing, (4) primer extension.
Table 1. Deoxynucleotides (dNTPs) for use in PCR, real-time PCR, and reverse transcription PCR
One-Step RT-PCR
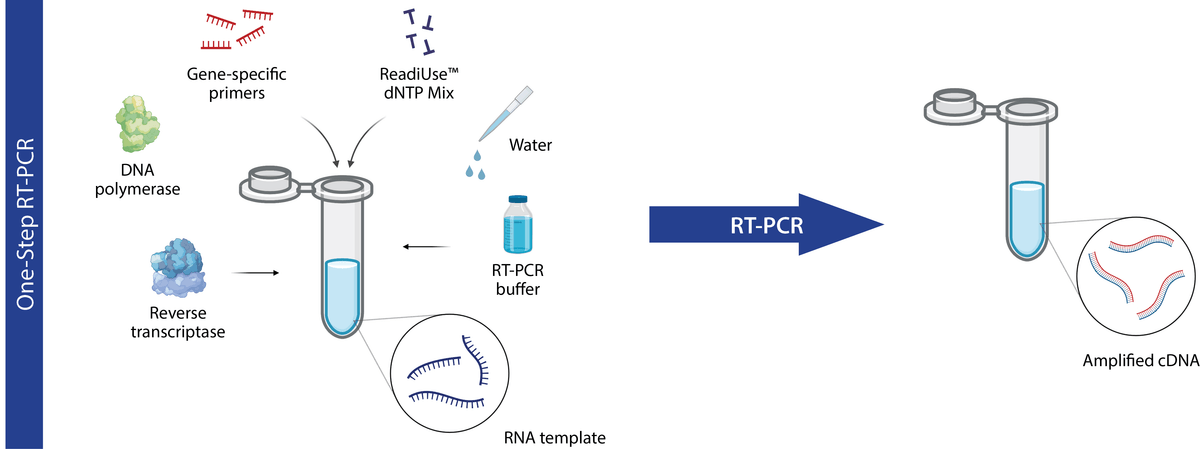
In one-step RT-PCR, reverse transcription (i.e. cDNA synthesis) and PCR are carried out in the same reaction vessel and buffer. This simple and convenient single-tube design works well when performing a small number of assays and is amenable to high-speed, high-throughput applications. In one-step RT-PCR, sequence-specific primers are required to direct both the synthesis and amplification of cDNA. Since specific primers more readily anneal at higher reaction temperatures than random primers, it is best to utilize reverse transcriptases with high thermal stability when using this approach.
One-step RT-PCR diagram. In one-step RT-PCR, cDNA synthesis via reverse transcription (RT) and subsequent PCR amplification occur in the same reaction vessel (figure made in BioRender).
While the one-step RT-PCR approach offers several advantages, it is not without its caveats. Since both reverse transcription and PCR take place in the same tube the reaction conditions cannot be separately optimized, which can impact yield or reaction efficiency to varying degrees. Moreover, because all the cDNA synthesized is consumed in the subsequent PCR step, additional aliquots of the original RNA sample(s) are required in order to repeat reactions or to assess the expression of other genes.
Advantages, disadvantages and applications for one-step RT-PCR
| One-Step RT-PCR | |
| RT Primers |
|
| Advantages |
|
| Disadvantages |
|
| Ideal Applications |
|
Two-Step RT-PCR
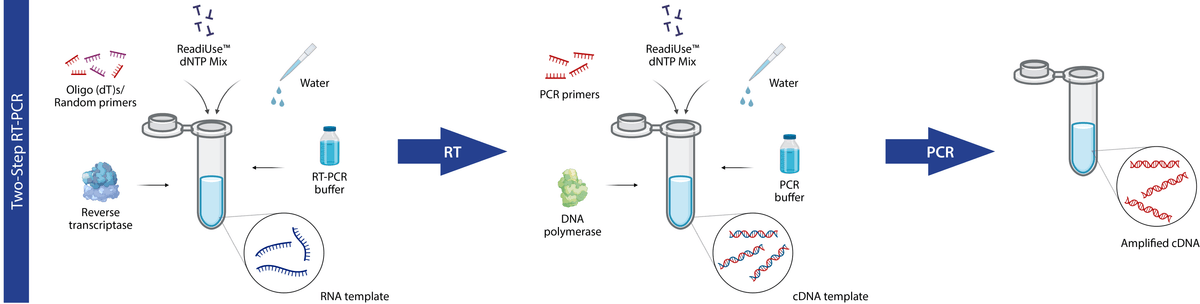
In two-step RT-PCR, reverse transcription and PCR are carried out in separate reaction vessels with different buffers, reaction conditions and priming strategies. Rather than employing only gene-specific primers like the one-step method, two-step RT-PCR is carried out using a mixture of random hexamers, oligo-dT primers, and/or gene-specific primers, which provides a full cDNA archive of all the RNA species in the sample. Furthermore, the uncoupling of reverse transcription and PCR allows for each reaction to be optimized individually for maximum efficiency and sequence representation. This permits a higher yield of cDNA during reverse transcription and provides an opportunity for cDNA samples to be stored for future amplification reactions or downstream applications.
Two-step RT-PCR diagram. In two-step RT-PCR, cDNA synthesis via reverse transcription (RT) and subsequent PCR amplification occur in separate reaction vessels (figure made in BioRender).
While two-step RT-PCR is highly sensitive it is significantly more time-consuming and less amenable to high-speed, high-throughput application than one-step RT-PCR. It requires an additional open-tube step, more pipetting and sample handling, which can increase variability and the risk of contamination.
Advantages, disadvantages and applications for two-step RT-PCR
| Two-Step RT-PCR | |
| RT Primers |
|
| Advantages |
|
| Disadvantages |
|
| Ideal Applications |
|
Measuring RT-PCR Amplicons
There are two primary approaches for measuring amplicon generation in RT-PCR. The first approach analyzes amplicon amplification after all the cycles of PCR have been completed and is referred to as end-point RT-PCR. The second approach measures amplicon concentration in real-time as it occurs throughout the PCR cycling process and is known as real-time RT-PCR or quantitative RT-PCR (RT-qPCR). In RT-qPCR, several different types of chemistries can be used to detect amplicon generation in real-time, these include Helixyte™ Green, Molecular Beacons and TaqMan probes.
End-point RT-PCR
End-point RT-PCR analysis, which is based on the plateau phase of PCR reaction, is used to analyze amplified products after all the cycles of the PCR reaction have been completed. In this method, amplicons are separated by agarose gel electrophoresis and visualized using a DNA binding dye, such as ethidium bromide (EtBr), to determine the size of the DNA molecules in the range of 500 to 30,000 bp. While EtBr is the most commonly used dye for visualizing DNA, it is mutagenic and highly toxic through inhalation. Instead, consider using safer and more environmentally friendly, non-toxic alternatives such as Gelite™ X100 (Cat No. 17706), Helixyte™ Green (Cat No. 17590), Helixyte™ Gold (Cat No. 17595), Gelite™ Green (Cat No. 17589) or Gelite™ Orange (Cat No. 17594).
Advantages, disadvantages and applications of end-point RT-PCR
| End-Point RT-PCR | |
| Advantages |
|
| Disadvantages |
|
| Applications |
|
Table 2. Nucleic acid stains for agarose and polyacrylamide gel electrophoresis
| Product ▲ ▼ | Ex (nm)¹ ▲ ▼ | Filter² ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| Helixyte™ Green Nucleic Acid Gel Stain *10,000X DMSO Solution* | 254 mn | Long path green filter | 1 mL | 17590 |
| Helixyte™ Green Nucleic Acid Gel Stain *10,000X DMSO Solution* | 254 mn | Long path green filter | 100 µL | 17604 |
| Helixyte™ Gold Nucleic Acid Gel Stain *10,000X DMSO Solution* | 254 mn | Long path green filter | 1 mL | 17595 |
| Gelite™ Green Nucleic Acid Gel Staining Kit | 254 nm or 300 nm | Long path green filter | 1 Kit | 17589 |
| Gelite™ Orange Nucleic Acid Gel Staining Kit | 254 nm or 300 nm | Long path green filter | 1 Kit | 17594 |
| Gelite™ Safe DNA Gel Stain *10,000X Water Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 100 µL | 17700 |
| Gelite™ Safe DNA Gel Stain *10,000X Water Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 500 µL | 17701 |
| Gelite™ Safe DNA Gel Stain *10,000X Water Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 1 mL | 17702 |
| Gelite™ Safe DNA Gel Stain *10,000X Water Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 10 mL | 17703 |
| Gelite™ Safe DNA Gel Stain *10,000X DMSO Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 100 µL | 17704 |
Quantitative RT-PCR
In quantitative RT-PCR (RT-qPCR), fluorescent DNA-intercalating dyes or sequence-specific fluorescent probes are integrated into the RT-PCR reaction allowing for amplicon concentration to be measured in real-time during the exponential phase of PCR. By combining amplification and detection into a single-step, RT-qPCR provides greater precision and accuracy and produces quantitative data with a dynamic range several orders of magnitude larger than end-point RT-PCR. Because of its higher sensitivity, RT-qPCR is routinely used to analyze mRNA in gene expression, to examine the presence of retroviruses and to validate results obtained by array analyses.
Helixyte™ Green for RT-qPCR
RT-qPCR using fluorescent DNA-intercalating dyes, such as Helixyte™ Green (Cat No.17591) provides the easiest and most economical method for detecting and quantitating PCR amplicons in real-time. Helixyte™ Green binds to double-stranded DNA (dsDNA), and when excited emits light. As amplicon concentration increases with each successive cycle of amplification, so does the fluorescence intensity of Helixyte™ Green, to a degree proportional to the amount of dsDNA present in each PCR cycle. Helixyte™ Green is a much safer alternative than the highly mutagenic EtBr and can be used to monitor the amplification of any dsDNA sequence with greater sensitivity and less PCR inhibition. Because DNA-intercalating dyes will bind to any dsDNA, such as primer-dimers and non-specific products, it is important to use well-designed primers to avoid amplifying non-target sequences. To ensure amplification specificity and to check for primer-dimer artifacts, a melt curve analysis should be performed post-amplification.
Table 3. Double-stranded DNA-binding dyes for qPCR
| Product ▲ ▼ | Ex (nm) ▲ ▼ | Em (nm) ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| Helixyte™ Green *20X Aqueous PCR Solution* | 498 nm | 522 nm | 5x1 mL | 17591 |
| Helixyte™ Green *10,000X Aqueous PCR Solution* | 498 nm | 522 nm | 1 mL | 17592 |
| Helixyte™ Green dsDNA Quantifying Reagent *200X DMSO Solution* | 490 nm | 525 nm | 1 mL | 17597 |
| Helixyte™ Green dsDNA Quantifying Reagent *200X DMSO Solution* | 490 nm | 525 nm | 10 mL | 17598 |
| Q4ever™ Green *1250X DMSO Solution* | 503 nm | 527 nm | 100 µL | 17608 |
| Q4ever™ Green *1250X DMSO Solution* | 503 nm | 527 nm | 2 mL | 17609 |
TaqMan® Probes and Molecular Beacons for RT-qPCR
In probe-based RT-qPCR, fluorescently-labeled, target-specific probes are used to measure DNA amplification in real-time. This method benefits from extreme specificity and affords the end-user the opportunity for multiplexing multiple targets in a single reaction. Of the many probe-based RT-qPCR chemistries available, TaqMan® probes and Molecular Beacons, are the most widely used and both depend upon Förster Resonance Energy Transfer (FRET) to generate a fluorescence signal. TaqMan® probes rely on the 5'-nuclease activity of Taq DNA polymerase. Short oligonucleotide sequences, complementary to the target of interest, are labeled with a fluorescent reporter dye at the 5' end (see Table 4 below) and a non-fluorescent quencher dye at the 3' end (see Table 5 below). During PCR cycling, primers and probe anneal to the target. As Taq DNA polymerase binds to and extends the primer upstream of the probe, the hybridized probe is hydrolyzed and the fragment containing the reporter dye is released. The fluorescence signal can now be detected and the amount of fluorescence signal generated is proportional to the amount of qPCR products produced.
Like TaqMan® probes, Molecular Beacons are labeled with a fluorescent reporter dye at the 5' end and a non-fluorescent quencher dye at the 3' end. However, this method does not rely on the 5' nuclease activity of Taq DNA polymerase to generate a signal, rather Molecular Beacons are designed to remain intact during the entire amplification process. In the absence of the target, Molecular Beacons remain in a 'hairpin' confirmation due to its self-complementary stem structure. This brings both the fluorescent reporter and quencher dyes within close proximity of one another preventing the probe from fluorescing. When the Molecular Beacon hybridizes to its target, the fluorescent reporter and the quencher are separated, and the reporter dye emits at its characteristic wavelength.
Table 4. Fluorescent reporter dyes for labeling the 5' end or 3' end on sequence-specific qPCR probes.
| Product ▲ ▼ | Ex (nm) ▲ ▼ | Em (nm) ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| EDANS acid [5-((2-Aminoethyl)amino)naphthalene-1-sulfonic acid] *CAS 50402-56-7* | 336 | 455 | 1 g | 610 |
| EDANS acid [5-((2-Aminoethyl)amino)naphthalene-1-sulfonic acid] *CAS 50402-56-7* | 336 | 455 | 10 g | 611 |
| EDANS C5 maleimide | 336 | 455 | 5 mg | 619 |
| EDANS sodium salt [5-((2-Aminoethyl)aminonaphthalene-1-sulfonic acid, sodium salt] *CAS 100900-07-0* | 336 | 455 | 1 g | 615 |
| EDANS sodium salt [5-((2-Aminoethyl)aminonaphthalene-1-sulfonic acid, sodium salt] *CAS 100900-07-0* | 336 | 455 | 10 g | 616 |
| Tide Fluor™ 1 acid [TF1 acid] *Superior replacement for EDANS* | 341 | 448 | 100 mg | 2238 |
| Tide Fluor™ 1 alkyne [TF1 alkyne] | 341 | 448 | 5 mg | 2237 |
| Tide Fluor™ 1 amine [TF1 amine] *Superior replacement for EDANS* | 341 | 448 | 5 mg | 2239 |
| Tide Fluor™ 1 azide [TF1 azide] | 341 | 448 | 5 mg | 2236 |
| Tide Fluor™ 1 CPG [TF1 CPG] *500 Å* | 341 | 448 | 100 mg | 2240 |
Table 5. Quencher dyes for labeling the 5' end or 3' end on sequence-specific qPCR probes.
| Product ▲ ▼ | Ex (nm) ▲ ▼ | Em (nm) ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| DABCYL acid [4-((4-(Dimethylamino)phenyl)azo)benzoic acid] *CAS 6268-49-1* | 454 | N/A | 5 g | 2001 |
| DABCYL C2 amine | 454 | N/A | 100 mg | 2006 |
| DABCYL C2 maleimide | 454 | N/A | 25 mg | 2008 |
| DABCYL-DBCO | 454 | N/A | 5 mg | 2010 |
| DABCYL succinimidyl ester [4-((4-(Dimethylamino)phenyl)azo)benzoic acid, succinimidyl ester] *CAS 146998-31-4* | 454 | N/A | 1 g | 2004 |
| DABCYL succinimidyl ester [4-((4-(Dimethylamino)phenyl)azo)benzoic acid, succinimidyl ester] *CAS 146998-31-4* | 454 | N/A | 5 g | 2005 |
| 3'-DABCYL CPG *1000 Å* | 454 | N/A | 1 g | 6008 |
| 5'-DABCYL C6 Phosphoramidite | 454 | N/A | 1 g | 6009 |
| Tide Quencher™ 1 acid [TQ1 acid] | 492 | N/A | 100 mg | 2190 |
| Tide Quencher™ 1 alkyne [TQ1 alkyne] | 492 | N/A | 5 mg | 2189 |
Table 6. Recommended FRET pairs for developing FRET oligonucleotides
| Donor \ Acceptor ▲ ▼ | DABCYL ▲ ▼ | TQ1 ▲ ▼ | TQ2 ▲ ▼ | TQ3 ▲ ▼ | TQ4 ▲ ▼ | TQ5 ▲ ▼ | TQ6 ▲ ▼ | TQ7 ▲ ▼ |
| EDANS | +++ | +++ | + | - | - | - | - | - |
| MCA | +++ | +++ | + | - | - | - | - | - |
| Tide Fluor™ 1 | +++ | +++ | + | - | - | - | - | - |
| FAM FITC | + | + | +++ | + | - | - | - | - |
| Cy2® Tide Fluor™ 2 | + | + | +++ | + | - | - | - | - |
| HEX JOE TET | - | - | + | +++ | + | - | - | - |
| Cy3® TAMRA Tide Fluor™ 3 | - | - | + | +++ | + | - | - | - |
| ROX Texas Red® | - | - | - | + | +++ | + | - | - |
| Tide Fluor™ 4 | - | - | - | + | +++ | + | - | - |
| Cy5® Tide Fluor™ 5 | - | - | - | - | + | +++ | + | - |