PCR Detection of Viral DNA/RNA
In viral diagnostics, nucleic acid amplification tests, such as reverse transcription polymerase chain reaction (RT-PCR), permits the early detection and identification of infectious diseases known as retroviruses. In this modified version of the standard PCR process, mRNA, which serves as the initial template, is first reversed transcribed to complementary DNA (cDNA) and then subsequently amplified via PCR for downstream analysis. Relative to other techniques for measuring mRNA, such as Northern blot analysis, RNAse protection assays, or in situ hybridization, RT-PCR is significantly more robust at detecting the RNA transcript of any gene regardless of its relative abundance. RT-PCR has become an instrumental diagnostic tool for the detection of pathogens, including influenza viruses, enteroviruses, coronavirus (SARS-CoV-2), Ebola virus, and HIV.
Retroviruses
A retrovirus is a type of virus whose genetic material is encoded in RNA. Similar to other viruses, retroviruses hijack the cell's machinery in order to replicate copies of themselves. Upon infecting a cell, the retrovirus utilizes its own reverse-transcriptase enzyme to generate double-stranded DNA from its RNA genome. The newly synthesized viral DNA is integrated into the host cell genome, and the virus thereafter replicates as part of the host cell's DNA.
In humans, many retroviruses cause serious diseases. Retroviruses such as HIV-1 and HIV-2 cause the disease AIDS, while the human T-lymphotropic virus causes leukemia (i.e. cancer of the blood) and demyelinating disease. The severity of such diseases and the constant threat of new emerging infectious diseases has reaffirmed the need for developing rapid diagnostic tools capable of detecting low-density infections. One of the most well-established tools for the early detection of retroviral infection is nucleic acid amplification testing.
Nucleic Acid Amplification Tests
A nucleic acid amplification test is designed to detect small amounts of a particular nucleic acid sequence in order to identify a specific pathogen, often a virus or bacteria. Rather than detecting antigens or antibodies (the objective in serological analysis), nucleic acid amplification tests target and detect genetic materials, such as DNA or RNA. The detection of genetic materials allows for early diagnosis of infection prior to seroconversion, the time period during which a specific antibody develops against the pathogen and becomes detectable in the blood. Common nucleic acid amplification tests include reverse transcription polymerase chain reaction (RT-PCR), strand displacement assay (SDA), or transcription mediated assay (TMA).
Reverse Transcription PCR (RT-PCR)
Reverse transcription polymerase chain reaction (RT-PCR) is a highly sensitive technique for the detection and quantitation of mRNA expression levels. In RT-PCR, the RNA template is first reversed transcribed into complementary DNA (cDNA), using the enzyme reverse transcriptase. This RNA-dependent DNA polymerase, with the assistance of reverse transcription primers, deoxynucleotides (dNTPs: dATP, dTTP, dGTP and dCTP), catalyzes the synthesis of cDNA from its respective target RNA sequence. The cDNA is then used as a template for exponential amplification using standard PCR procedure (denaturation, annealing and elongation). RT-PCR is used in a variety of applications including gene expression analysis, microarray validation, pathogen detection, and disease research.
Quantification of RT-PCR products can be generally divided into two categories: end-point RT-PCR and quantitative RT-PCR (RT-qPCR).
Table 1. Deoxynucleotides (dNTPs) for use in PCR, real-time PCR, and reverse transcription PCR
End-point RT-PCR
End-point RT-PCR analysis, which is based on the plateau phase of PCR reaction, is used to analyze amplified products after all the cycles of the PCR reaction have been completed. In this method, amplicons are separated by agarose gel electrophoresis and visualized using a DNA binding dye, such as ethidium bromide (EtBr), to determine the size of the DNA molecules in the range of 500 to 30,000 bp. While EtBr is the most commonly used dye for visualizing DNA, it is mutagenic and highly toxic through inhalation. Instead, consider using safer and more environmentally friendly, non-toxic alternatives such as Gelite™ X100 (Cat No. 17706), Helixyte™ Green (Cat No. 17590), Helixyte™ Gold (Cat No. 17595), Gelite™ Green (Cat No. 17589) or Gelite™ Orange (Cat No. 17594).
Table 2. End-point RT-PCR detection methods For Quantifying RT-PCR products
| End-Point RT-PCR ▲ ▼ | Overview ▲ ▼ |
| Relative RT-PCR |
|
| Competitive RT-PCR |
|
| Comparative RT-PCR |
|
Table 3. Advantages, disadvantages and applications of end-point RT-PCR
▲ ▼ | End-Point RT-PCR ▲ ▼ |
| Advantages |
|
| Disadvantages |
|
| Applications |
|
Table 4. Nucleic acid stains for agarose and polyacrylamide gel electrophoresis
| Product ▲ ▼ | Ex (nm)¹ ▲ ▼ | Filter² ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| Helixyte™ Green Nucleic Acid Gel Stain *10,000X DMSO Solution* | 254 mn | Long path green filter | 1 mL | 17590 |
| Helixyte™ Green Nucleic Acid Gel Stain *10,000X DMSO Solution* | 254 mn | Long path green filter | 100 µL | 17604 |
| Helixyte™ Gold Nucleic Acid Gel Stain *10,000X DMSO Solution* | 254 mn | Long path green filter | 1 mL | 17595 |
| Gelite™ Green Nucleic Acid Gel Staining Kit | 254 nm or 300 nm | Long path green filter | 1 Kit | 17589 |
| Gelite™ Orange Nucleic Acid Gel Staining Kit | 254 nm or 300 nm | Long path green filter | 1 Kit | 17594 |
| Gelite™ Safe DNA Gel Stain *10,000X Water Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 100 µL | 17700 |
| Gelite™ Safe DNA Gel Stain *10,000X Water Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 500 µL | 17701 |
| Gelite™ Safe DNA Gel Stain *10,000X Water Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 1 mL | 17702 |
| Gelite™ Safe DNA Gel Stain *10,000X Water Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 10 mL | 17703 |
| Gelite™ Safe DNA Gel Stain *10,000X DMSO Solution* | 254 nm, 300 nm or 520 nm | Ethidium Bromide, Gel Star, Gel Green, Gel Red and SYBR filters | 100 µL | 17704 |
Quantitative RT-PCR
In quantitative RT-PCR (RT-qPCR), fluorescent DNA-intercalating dyes or sequence-specific fluorescent probes are integrated into the RT-PCR reaction allowing for amplicon concentration to be measured in real-time during the exponential phase of PCR. By combining amplification and detection into a single-step, RT-qPCR provides greater precision and accuracy and produces quantitative data with a dynamic range several orders of magnitude larger than end-point RT-PCR. Because of its higher sensitivity, RT-qPCR is routinely used to analyze mRNA in gene expression, to examine the presence of retroviruses and to validate results obtained by array analyses.
Helixyte™ Green for RT-qPCR
RT-qPCR using fluorescent DNA-intercalating dyes, such as Helixyte™ Green (Cat No.17591) provides the easiest and most economical method for detecting and quantitating PCR amplicons in real-time. Helixyte™ Green binds to double-stranded DNA (dsDNA), and when excited emits light. As amplicon concentration increases with each successive cycle of amplification, so does the fluorescence intensity of Helixyte™ Green, to a degree proportional to the amount of dsDNA present in each PCR cycle. Helixyte™ Green is a much safer alternative than the highly mutagenic EtBr and can be used to monitor the amplification of any dsDNA sequence with greater sensitivity and less PCR inhibition. Because DNA-intercalating dyes will bind to any dsDNA, such as primer-dimers and non-specific products, it is important to use well-designed primers to avoid amplifying non-target sequences. To ensure amplification specificity and to check for primer-dimer artifacts, a melt curve analysis should be performed post-amplification.
Table 5. Double-stranded DNA-binding dyes for qPCR
| Product ▲ ▼ | Ex (nm) ▲ ▼ | Em (nm) ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| Helixyte™ Green *20X Aqueous PCR Solution* | 498 nm | 522 nm | 5x1 mL | 17591 |
| Helixyte™ Green *10,000X Aqueous PCR Solution* | 498 nm | 522 nm | 1 mL | 17592 |
| Helixyte™ Green dsDNA Quantifying Reagent *200X DMSO Solution* | 490 nm | 525 nm | 1 mL | 17597 |
| Helixyte™ Green dsDNA Quantifying Reagent *200X DMSO Solution* | 490 nm | 525 nm | 10 mL | 17598 |
| Q4ever™ Green *1250X DMSO Solution* | 503 nm | 527 nm | 100 µL | 17608 |
| Q4ever™ Green *1250X DMSO Solution* | 503 nm | 527 nm | 2 mL | 17609 |
TaqMan® Probes and Molecular Beacons for RT-qPCR
In probe-based RT-qPCR, fluorescently-labeled, target-specific probes are used to measure DNA amplification in real-time. This method benefits from extreme specificity and affords the end-user the opportunity for multiplexing multiple targets in a single reaction. Of the many probe-based RT-qPCR chemistries available, TaqMan® probes and Molecular Beacons, are the most widely used and both depend upon Förster Resonance Energy Transfer (FRET) to generate a fluorescence signal. TaqMan® probes rely on the 5'-nuclease activity of Taq DNA polymerase. Short oligonucleotide sequences, complementary to the target of interest, are labeled with a fluorescent reporter dye at the 5' end (see Table 5 below) and a non-fluorescent quencher dye at the 3' end (see Table 6 below). During PCR cycling, primers and probe anneal to the target. As Taq DNA polymerase binds to and extends the primer upstream of the probe, the hybridized probe is hydrolyzed and the fragment containing the reporter dye is released. The fluorescence signal can now be detected and the amount of fluorescence signal generated is proportional to the amount of qPCR products produced.
Like TaqMan® probes, Molecular Beacons are labeled with a fluorescent reporter dye at the 5' end and a non-fluorescent quencher dye at the 3' end. However, this method does not rely on the 5' nuclease activity of Taq DNA polymerase to generate a signal, rather Molecular Beacons are designed to remain intact during the entire amplification process. In the absence of the target, Molecular Beacons remain in a 'hairpin' confirmation due to its self-complementary stem structure. This brings both the fluorescent reporter and quencher dyes within close proximity of one another preventing the probe from fluorescing. When the Molecular Beacon hybridizes to its target, the fluorescent reporter and the quencher are separated, and the reporter dye emits at its characteristic wavelength.
We offer a broad range of dye phosphoramidites and dye CPG supports for developing FRET oligonucleotides, TaqMan® probes and Molecular Beacons. This set includes classic dyes such as FAM, HEX, TET and JOE, as well as superior oligo-labeling dyes such as Tide Fluor™ and Tide Quencher™ dyes.
Table 6. Fluorescent reporter dyes for labeling the 5' end or 3' end on sequence-specific qPCR probes.
| Product ▲ ▼ | Ex (nm) ▲ ▼ | Em (nm) ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| EDANS acid [5-((2-Aminoethyl)amino)naphthalene-1-sulfonic acid] *CAS 50402-56-7* | 336 | 455 | 1 g | 610 |
| EDANS acid [5-((2-Aminoethyl)amino)naphthalene-1-sulfonic acid] *CAS 50402-56-7* | 336 | 455 | 10 g | 611 |
| EDANS C5 maleimide | 336 | 455 | 5 mg | 619 |
| EDANS sodium salt [5-((2-Aminoethyl)aminonaphthalene-1-sulfonic acid, sodium salt] *CAS 100900-07-0* | 336 | 455 | 1 g | 615 |
| EDANS sodium salt [5-((2-Aminoethyl)aminonaphthalene-1-sulfonic acid, sodium salt] *CAS 100900-07-0* | 336 | 455 | 10 g | 616 |
| Tide Fluor™ 1 acid [TF1 acid] *Superior replacement for EDANS* | 341 | 448 | 100 mg | 2238 |
| Tide Fluor™ 1 alkyne [TF1 alkyne] | 341 | 448 | 5 mg | 2237 |
| Tide Fluor™ 1 amine [TF1 amine] *Superior replacement for EDANS* | 341 | 448 | 5 mg | 2239 |
| Tide Fluor™ 1 azide [TF1 azide] | 341 | 448 | 5 mg | 2236 |
| Tide Fluor™ 1 CPG [TF1 CPG] *500 Å* | 341 | 448 | 100 mg | 2240 |
Table 7. Quencher dyes for labeling the 5' end or 3' end on sequence-specific qPCR probes.
| Product ▲ ▼ | Ex (nm) ▲ ▼ | Em (nm) ▲ ▼ | Unit Size ▲ ▼ | Cat No. ▲ ▼ |
| DABCYL acid [4-((4-(Dimethylamino)phenyl)azo)benzoic acid] *CAS 6268-49-1* | 454 | N/A | 5 g | 2001 |
| DABCYL C2 amine | 454 | N/A | 100 mg | 2006 |
| DABCYL C2 maleimide | 454 | N/A | 25 mg | 2008 |
| DABCYL-DBCO | 454 | N/A | 5 mg | 2010 |
| DABCYL succinimidyl ester [4-((4-(Dimethylamino)phenyl)azo)benzoic acid, succinimidyl ester] *CAS 146998-31-4* | 454 | N/A | 1 g | 2004 |
| DABCYL succinimidyl ester [4-((4-(Dimethylamino)phenyl)azo)benzoic acid, succinimidyl ester] *CAS 146998-31-4* | 454 | N/A | 5 g | 2005 |
| 3'-DABCYL CPG *1000 Å* | 454 | N/A | 1 g | 6008 |
| 5'-DABCYL C6 Phosphoramidite | 454 | N/A | 1 g | 6009 |
| Tide Quencher™ 1 acid [TQ1 acid] | 492 | N/A | 100 mg | 2190 |
| Tide Quencher™ 1 alkyne [TQ1 alkyne] | 492 | N/A | 5 mg | 2189 |
Table 8. Recommended FRET pairs for developing FRET oligonucleotides
| Donor \ Acceptor ▲ ▼ | DABCYL ▲ ▼ | TQ1 ▲ ▼ | TQ2 ▲ ▼ | TQ3 ▲ ▼ | TQ4 ▲ ▼ | TQ5 ▲ ▼ | TQ6 ▲ ▼ | TQ7 ▲ ▼ |
| EDANS | +++ | +++ | + | - | - | - | - | - |
| MCA | +++ | +++ | + | - | - | - | - | - |
| Tide Fluor™ 1 | +++ | +++ | + | - | - | - | - | - |
| FAM FITC | + | + | +++ | + | - | - | - | - |
| Cy2® Tide Fluor™ 2 | + | + | +++ | + | - | - | - | - |
| HEX JOE TET | - | - | + | +++ | + | - | - | - |
| Cy3® TAMRA Tide Fluor™ 3 | - | - | + | +++ | + | - | - | - |
| ROX Texas Red® | - | - | - | + | +++ | + | - | - |
| Tide Fluor™ 4 | - | - | - | + | +++ | + | - | - |
| Cy5® Tide Fluor™ 5 | - | - | - | - | + | +++ | + | - |
Tide Fluor™ Dyes for Labeling Oligos and Peptides
Tide Fluor™ dyes are a series of donor dyes optimized for developing FRET oligonucleotides and peptides for a variety of biological applications. Compared to common donor dyes such as EDANS, FAM, TAMRA, ROX, Cy 3 and Cy5, Tide Fluor™ dyes exhibit stronger fluorescence and higher photostability. They are the best affordable fluorescent dyes for labeling peptides and oligonucleotides without sacrificing performance.
Learn More
Table 9. Tide Fluor™ Dyes and Spectral Properties For Quantifying RT-PCR products
| Labeling Dye ▲ ▼ | Abs (nm) ▲ ▼ | Em (nm) ▲ ▼ | ε1 ▲ ▼ | Φ2 ▲ ▼ | CF at 260 nm3 ▲ ▼ | CF at 280 nm4 ▲ ▼ |
| Tide Fluor™ 1 | 345 | 442 | 20000 | 0.95 | 0.246 | 0.187 |
| Tide Fluor™ 2WS | 502 | 525 | 75000 | 0.9 | 0.211 | 0.091 |
| Tide Fluor™ 2 | 500 | 527 | 75000 | 0.9 | 0.288 | 0.201 |
| Tide Fluor™ 3WS | 555 | 565 | 150000 | 0.105 | 0.079 | 0.079 |
| Tide Fluor™ 3 | 555 | 584 | 85000 | 0.85 | 0.331 | 0.201 |
| Tide Fluor™ 4 | 590 | 618 | 90000 | 0.91 | 0.489 | 0.436 |
| Tide Fluor™ 5WS | 649 | 664 | 250000 | 0.25 | 0.023 | 0.027 |
| Tide Fluor™ 6WS | 676 | 695 | 220000 | 0.18 | 0.111 | 0.009 |
| Tide Fluor™ 7WS | 749 | 775 | 275000 | 0.12 | 0.009 | 0.049 |
| Tide Fluor™ 8WS | 775 | 807 | 250000 | 0.08 | 0.103 | 0.109 |
- ε = molar extinction coefficient at their maximum absorption wavelength (Units = cm-1M-1).
- Φ = Fluorescence quantum yield in aqueous buffer (pH 7.2).
- CF at 260 nm is the correction factor used for eliminating the dye contribution to the absorbance at 260 nm (for oligos and nucleic acid labeling).
- CF at 280 nm is the correction factor used for eliminating the dye contribution to the absorbance at 280 nm (for peptide and protein labeling).
Tide Quencher™ Dyes for Labeling Oligos and Peptides
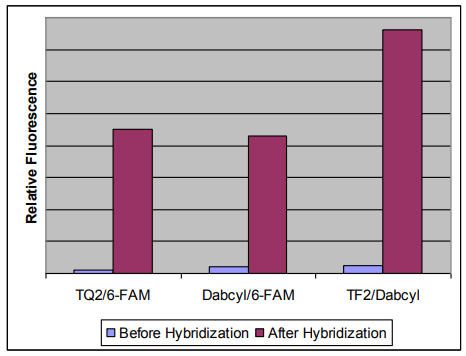
Hybridization-induced fluorescence enhancement of Molecular Beacon oligonucleotide probes that contain Tide Fluor™ dyes as donor or Tide Quencher™ dyes as acceptors.
The Key Benefits of Tide Quencher™ Dyes:
- Explore the FRET potentials that might be impossible with other quenchers.
- Versatile reactive forms are convenient for self-constructing your desired FRET biomolecules.
- Perfectly match your desired fluorescent donors.
- Competitive price with better performance.
Table 10. Tide Quencher™ Dyes and Spectral Properties For Quantifying RT-PCR products
| Quencher ▲ ▼ | Abs (nm) ▲ ▼ | ε*1 ▲ ▼ | CF at 260 nm*2 ▲ ▼ | CF at 280 nm*3 ▲ ▼ |
| Tide Quencher™ 1 | 488 | 20,000 | 0.147 | 0.194 |
| Tide Quencher™ 2 | 512 | 21,000 | 0.100 | 0.120 |
| Tide Quencher™ 2WS | 515 | 21,000 | 0.100 | 0.120 |
| Tide Quencher™ 3 | 576 | 22,000 | 0.085 | 0.091 |
| Tide Quencher™ 3WS | 576 | 90,000 | 0.186 | 0.205 |
| Tide Quencher™ 4 | 604 | 23,000 | 0.146 | 0.183 |
| Tide Quencher™ 4WS | 604 | 90,000 | 0.149 | 0.136 |
| Tide Quencher™ 5 | 661 | 24,000 | 0.170 | 0.082 |
| Tide Quencher™ 5WS | 661 | 130,000 | 0.072 | 0.082 |
| Tide Quencher™ 6WS | 691 | 130,000 | 0.120 | 0.102 |
- ε = molar extinction coefficient at their maximum absorption wavelength (Units = cm-1M-1).
- CF at 260 nm is the correction factor used for eliminating the dye contribution to the absorbance at 260 nm (for oligos and nucleic acid labeling).
- CF at 280 nm is the correction factor used for eliminating the dye contribution to the absorbance at 280 nm (for peptide and protein labeling).